Introduction
Recent years have seen an explosion in interest in the use of eDNA to detect the presence of individual species in the environment with targeted assays, as opposed to biodiversity assessments via metabarcoding. The number of published assays is increasing every year, while there is growing interest from policy makers in implementing the tests for decision-making in environmental management.
A challenge faced by potential users is the huge variation in the level to which published assays have been validated, making it difficult to work out whether or not an assay is reliable enough for application in contexts where expensive management interventions may be applied based on the results.
The 5-level validation scale presented here seeks to address this by providing a simple framework in which to place assays according to the level of validation they have achieved.
1. The validation scale

An overview of the 5-level validation scale. For each of the levels (incomplete to operational), the main accomplishments in the validation process and an appropriate interpretation of results are provided.
2. Variable blocks and minimum criteria
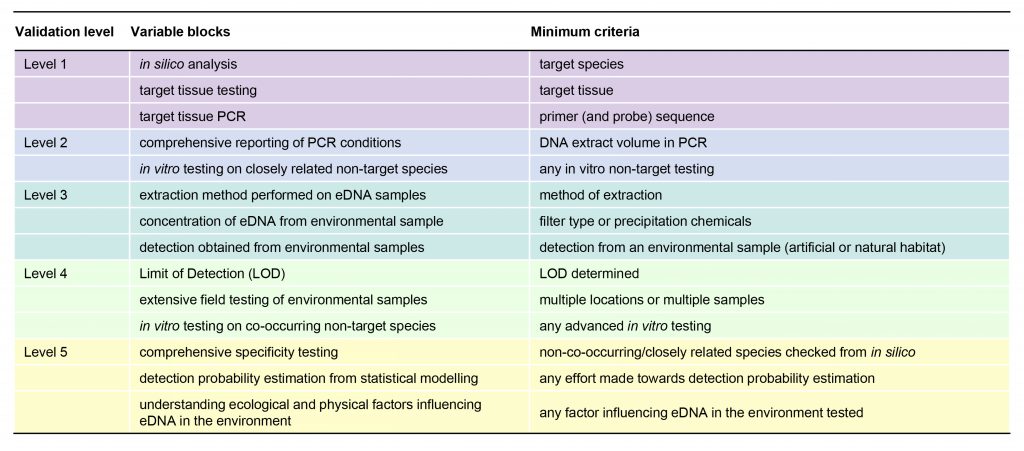
The thematic variable blocks of the 5-level validation scale and their respective minimum criteria (MC).
The MC provide a proof of validation as each block contains variables associated with either rudimentary or substantial validation and reporting. For an assay to be placed at a certain level of the validation scale, it must fulfil the MC of all variable blocks associated with this level. The scale is additive; hence, Level 2 assays must fulfil the Level 2 MC in addition to the Level 1 MC.
3. Remaining uncertainties at each level
| Level 1 | Level 2 | Level 3 | Level 4 | Level 5 |
|---|---|---|---|---|
| DNA may have been introduced by other organisms, including humans. If the target is also a food item then it could have been introduced via waste water | DNA may have been introduced by other organisms, including humans. If the target is also a food item then it could have been introduced via waste water | DNA may have been introduced by other organisms, including humans. If the target is also a food item then it could have been introduced via waste water | DNA may have been introduced by other organisms, including humans. If the target is also a food item then it could have been introduced via waste water | DNA may have been introduced by other organisms, including humans. If the target is also a food item then it could have been introduced via waste water |
| It is not known how ecological and seasonal factors may influence detectability. | It is not known how ecological and seasonal factors may influence detectability. | It is not known how ecological and seasonal factors may influence detectability. | It is not known how ecological and seasonal factors may influence detectability. | Some untested ecological factors may still influence detectability. |
| It is not known how many samples are required for a 95% chance of detection. | It is not known how many samples are required for a 95% chance of detection. | It is not known how many samples are required for a 95% chance of detection. | It is not known how many samples are required for a 95% chance of detection. | Sensitivity may vary in different environments. |
| Co-occurring species that have not been tested may co-amplify, leading to false positive results. PCR products should be sequenced to verify target presence. | Co-occurring species that have not been tested may co-amplify, leading to false positive results. PCR products should be sequenced to verify target presence. | Co-occurring species that have not been tested may co-amplify, leading to false positive results. PCR products should be sequenced to verify target presence. | Unrelated species may occasionally co-amplify, although the risk is low given the extensive field testing undertaken. Additional specificity testing may need to be undertaken if using the assay in a different region to that in which it was originally validated. | Unrelated species may occasionally co-amplify, although the risk is low given the extensive field testing undertaken. Additional specificity testing may need to be undertaken if using the assay in a different region to that in which it was originally validated. |
| It is not known if this assay will work on environmental samples. | It is not known if this assay will work on environmental samples. |
The content of this section can not be displayed in the mobile version of the page.
4. Valid interpretation of eDNA-based results
| Level 1 | Level 2 | Level 3 | Level 4 | Level 5 | |
|---|---|---|---|---|---|
| Negative Result | Impossible to tell if target is present or absent | Impossible to tell if target is present or absent | Impossible to tell if target is present or absent | Target is likely to be absent, assuming sampling has been carried out at an appropriate time of year and with a high level of replication | Target is likely to be absent. Assuming appropriate sampling has been carried out, a probability of species presence (false negative) can be given. |
| Positive Result – Amplification | Impossible to tell if target is present or absent | Impossible to tell if target is present or absent | Target is likely to be present | Target is very likely to be present | Target is very likely to be present |
| Positive Result – Sequenced* | DNA of target is definitely present | DNA of target is definitely present | DNA of target is definitely present | DNA of target is definitely present | DNA of target is definitely present |
*Contamination is always a possibility; therefore, enough negative controls need to be generated from field sampling to PCR along the whole processing chain. This is especially important if short target DNA fragments (below 100 base pairs) are amplified and the sequenced fragment consists mainly of primer sequences and does not allow for discrimination between species.
The content of this section can not be displayed in the mobile version of the page.
5. Published targeted eDNA assays and validation scores
The associated literature database originally contained 327 papers with eDNA assays published until April 2019 which target specific species. The assay validation database contains information on validation and reporting of 546 previously published targeted eDNA assays. The original versions of both databases form the basis for a meta-analysis carried out by Thalinger et al. (https://doi.org/10.1002/edn3.189). We want to encourage assay developers to use the validation scale and checklist as a template to check the validation status of both newly developed and previously published assays.
Validation scores of additional assays can be submitted at the assay validation tab.
If you are interested in the validation status of an assay currently not contained in the database or would like to report errors, please contact edna.validation@gmail.com.
A French version of the Validation scale and the minimum criteria was recently generated by Abbott et al. (2021).*
*Abbott, C., Coulson, M., Gagné, N., Lacoursière‐Roussel, A., Parent, G.J., Bajno, R., Dietrich, C., May-McNally, S. (2021) Guidance on the Use of Targeted Environmental DNA (eDNA) Analysis for the Management of Aquatic Invasive Species and Species at Risk. Research Document 2021/019, Fisheries and Oceans Canada, https://www.dfo-mpo.gc.ca/csas-sccs/Publications/ResDocs-DocRech/2021/2021_019-eng.html
A German version of the Validation scale and the minimum criteria was recently generated by Elvira Mächler (SimplexDNA).
We hope that when new assays are published, reference will be made to this scale as a standardised way to communicate the extent to which they have been validated. It will help end users to decide whether or not an assay is suitable for their needs in terms of its level of validation and reporting, and regarding the remaining uncertainties around positive or negative results. The validation scale can also guide next steps for the advancement of assays in order to move them further along the scale.
Please note that the validation scale functions as a general framework, and that future technological advances and knowledge gains can make changes in the structure necessary. The scale was originally developed at a DNAqua-net (Working Group 3: field and laboratory methods) workshop in Innsbruck, Austria, in March 2018. For a complete assessment of targeted eDNA validation and reporting practices, please see Thalinger et al. (https://doi.org/10.1101/2020.04.27.063990)
At present, no assay reaches the top level on the scale, but this does not necessarily mean that published assays to date don’t work well. It simply means that care should be taken in basing management responses purely on the results of a semi-validated assay, particularly in regulatory contexts. However, there are times when a semi-validated assay can still be a useful tool to gain an idea of whether particular species may or may not be present – especially when used in combination with other survey methodologies (themselves often imperfect). What is critical is that the user understands the assay is not fully validated and bears this in mind when interpreting results. Therefore, for each level on the scale, we also present the remaining uncertainties that users should be aware of, as well as guidance on how a positive or negative result should be interpreted. Importantly, we also include a set of key assumptions about how the analyses have been conducted.
The scale is based on 122 variables, 104 of which are binary variables and can either be 0 for “no, not done or not reported” or 1 for “done or reported”. Based on the current assay reporting practices in the literature, it is often not possible to distinguish between a variable that has not been addressed and a variable that has not been reported.
The 104 variables can be divided in 14 thematic blocks, such as “in silico analysis” or “extensive field testing of environmental samples”, which are associated with different levels of the validation scale. For example, “in silico analysis” is regarded as a very basic requirement and hence associated with Level 1 assays. “Extensive field testing of environmental samples” is considered an advanced part of the validation process and is thus associated with Level 4 assays.
Each of the variable blocks contains variables which are already considered standard in assay validation and reporting (e.g. report primer and probe sequences) and variables which today are hardly ever reported/validated but are likely to have substantial effect on the results (e.g. pressure used for filtration). As proof of a rudimentary validation effort, we defined minimum criteria (MC) for each variable block, e.g. “primer (and probe) sequence reported” is the minimum criterion for the “target tissue PCR” variable block. For an assay to be placed at a certain level of the validation scale, it must fulfil the MC of all variable blocks associated with this level. The scale is additive; hence, Level 2 assays must fulfil the Level 2 MC in addition to the Level 1 MC.
Note that the placement on the validation scale is location-specific, such that if an assay is to be used in a different region from the one where it was validated, additional field trials will need to be carried out in the new region to ensure the same level of performance is achieved. If additional closely related species are present in the new region, this warrants additional specificity testing.
The team behind the scale
Kat Bruce, NatureMetrics
Bettina Thalinger, University of Guelph
Kristy Deiner, Natural History Museum in London & ETH Zurich
Helen Rees, ADAS
Daniela Sint, University of Innsbruck & Sinsoma
Rosie Blackman, EAWAG & University of Zurich
Lynsey Harper, Liverpool John Moores University
Michael Traugott, University of Innsbruck & Sinsoma
Caren Goldberg, Washington State University











